

|
CSI-ANN allows the user to predict regulatory DNA elements using chromatin modification data. The user provides chromatin modification data (e.g., H3K4me1, H3K4me3, H3K27ac, etc), either ChIP-chip or ChIP-Seq, and the sotfware produces a text file containing the prediction centers of regulatory DNA elements (genomic positions) across the regions/genomes. |
|
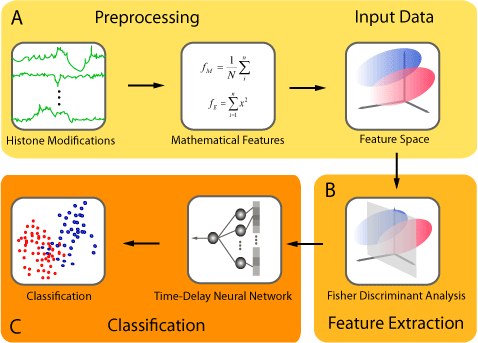
The CSI-ANN framework consists of a data transformation and a feature extraction step followed by a classification step using Time-Delay Neural Network (TDNN). Chromatin modification data can be used directly by the software (i.e. using summary statistic in a window) or processed through a pre-processing step to generate additional features (e.g., statistical measures such as variance, kustosis). Input Data are provided to the software in a prespecified format (see T cell sample files provided). The software is written in C. |
|
|
|
|
|
|
The CSI-ANN method is described in the following paper: Firpi HA, Ucar D, and Tan K. Discover regulatory DNA elements using chromatin signature and artificial neural network. 2010. Bioinformatics. 26(13):1579-1586. Email any questions, comments, or bugs found to Kai Tan (tank1@email.chop.edu) |