miPALM (Module Inference
by Parametric Local Modularity)
miPALM is a novel algorithm for detecting protein complexes from large protein protein interaction networks with improved accuracy than previous methods.
The algorithm uses a novel graph theoretic measure, parametric local modularity, to identify highly connected sub-networks as candidate protein complexes.
The software is implemented in Matlab
and is freely available for academic use.
Please cite: Jongkwang Kim and Kai Tan. Discover Protein Complexes in Protein-Protein Interaction Networks Using Parametric
Local Modularity BMC Bioinformatics 2010. 11:521.
Usage
miPALM -a 0.364 -d 2.4 [-i
input.net] [-o output.txt]
where -a and -d are standard parameters of miPALM and other command line parameters are optional:
-
-i (default standard dialog box) is a Pajek file containing unique node IDs and interactions among them.
-
-o (default standard file output) is the file to contain the detected complexes by miPALM. Default output file will be named as "input_a0.364_d2.4.txt" in "results" sub-folder.
Example
Given the input file called "my_input.net":
*Vertices 4
1 "P46677"
2 "P21372"
3 "P20433"
4 "Q00916"
*Edges
1 2
1 3
2 3
2 4
3 4
Please note that the example above has no edge-weight on the 3rd column and no vertex-position.
-
miPALM might be run as
miPALM -a 0.374 -d 2.33
-
To specify a user input file name (using the default output-file name)
miPALM -a 0.374 -d 2.33 -i my_input.net
-
To specify a user!/s output file name (using the default dialog box for input-file)
miPALM -a 0.374 -d 2.33 -o my_output.net
-
To input "Yeast_PPI_DIP_FULL.net" in the sub-folder "data" and to generate output file "my_output.txt" in the sub-folder "results"
miPALM -a 0.374 -d 2.33 -i data\Yeast_PPI_DIP_FULL.net -o results\my_output.txt
The generated "my_output.txt" looks like this:
Rank Score Proteins Complex
--------------------------------------------------------------
1 5.75 9 P09119 P54784 P32833 P54791
P54790 P38826 P50874 P32579 P38185
2 5.33 10 P41543
P39007 P33767 P48439 Q99380 Q92316 P46964 P53062 P47818 Q04629
3 5.25 9 P45976
Q06102 P29468 Q12102 Q01329 P38912 P39927 P53538 Q03713
4 5.09 12 P32074
P22804 P11076 P20606 P22214 P25385 P22213 P41834 P40509 P05738 P43621 P28791
5 5.00 9 P34253
Q04868 Q06706 P38874 Q02908 P42935 Q02884 Q06177 P46974
6 5.00 5 P36116
P39904 Q99260 P47061 Q12071
7 5.00 7 P40480
P36124 P14832 P25357 P53685 P53096 P22943
8 4.89 10 Q07468
P53129 P37898 P12868 Q03308 P27801 P39702 P38959 P20795 P38273
9 4.83 13 P09547
P32479 P32480 P22082 P18480 P43554 P32591 P38064 Q06090 P53628 Q05123 Q12406
Q06349
10 4.75 9 P32349
P22276 P35718 P32910 P04051 P20436 Q04307 P17890 P47076
In addition to *.txt file above, miPALM also generates another text file to contain protein node-numbers of a complex in each line. For more details, see "README.txt" file in the uncompressed folder.
For your subsequent analysis and visualization, miPALM automatically exports *.txt file to *.sif and *.net file for each predicted complex which can be imported into Cytoscape and Pajek, respectively.
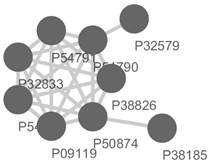
Figure 1. The top-scored sub-network in Yeast DIP-PPI
network detected by miPALM and visualized by Cytoscape.
Email to tank1@email.chop.edu for
bug-reporting.